Converting a dataset
This tutorial will guide you through the steps for the conversion of an existing dataset. We will use the VanDam-Daylong dataset from HomeBank as an example.
This is mainly an introduction to the way ChildProject handles things and how files are articulated together. If you need to create an entire new dataset, consider following our handbook guide, or making use of our ready to go datalad procedures to create the base structure and linkage to an online repository.
Set-up datalad and child-project
Make sure you have followed the Installation instructions before proceeding.
Create a dataset
The first step is to create a new dataset named vandam-data :
To do this, you will need to use a terminal and navigate to the folder you want your dataset created in.
Open your terminal or Anaconda prompt for Windows and activate your childproject environment with conda activate childproject
and use the cd command to navigate to your working directory.
This command will not only create the necessary folders but also the necessary datalad configuration files.
datalad create vandam-data
cd vandam-data
datalad create vandam-data
cd vandam-data
So far, the dataset contains nothing but hidden files:
ls -A
output:
.datalad .git .gitattributes
dir /a
output:
2011-01-01 04:32 PM <DIR> .
2011-01-01 04:32 PM <DIR> ..
2011-01-01 04:32 PM <DIR> .datalad
2011-01-01 04:32 PM <DIR> .git
2011-01-01 04:32 PM <DIR> .gitattributes
Now, we would like to get the data from https://homebank.talkbank.org/access/Public/VanDam-Daylong.html, convert it to our standards, and then publish it.
Gather and sort the files
The first step is to create the directories:
mkdir metadata # Create the metadata subfolder
mkdir -p recordings/raw # Create the subfolders for raw recordings
mkdir annotations # Create the subfolder for annotations
mkdir extra # Create the subfolder for extra data (that are neither metadata, recordings or annotations)
touch extra/.gitignore # Make sure the directory is present even though it's empty
mkdir metadata
mkdir recordings\raw
mkdir annotations
mkdir extra
type nul >> extra\.gitignore
Then, download the original data-set from HomeBank. You need to identify what key files are necessary for the dataset. We will get the audio files, the annotation files and some metadata.
The audio first:
curl https://media.talkbank.org/homebank/Public/VanDam-Daylong/BN32/BN32_010007.mp3 -o recordings/raw/BN32_010007.mp3
curl https://media.talkbank.org/homebank/Public/VanDam-Daylong/BN32/BN32_010007.mp3 -o recordings\raw\BN32_010007.mp3
Now let’s get the annotations.
curl https://homebank.talkbank.org/data/Public/VanDam-Daylong.zip -o VanDam-Daylong.zip
unzip VanDam-Daylong.zip
rm VanDam-Daylong.zip # Remove the zip archive
curl https://homebank.talkbank.org/data/Public/VanDam-Daylong.zip -o VanDam-Daylong.zip
tar -xf VanDam-Daylong.zip
del VanDam-Daylong.zip
Let’s explore the contents of VanDam-Daylong:
find . -not -path '*/\.*' -type f -print
output:
./recordings/raw/BN32_010007.mp3
./VanDam-Daylong/BN32/0its/e20100728_143446_003489.its
./VanDam-Daylong/BN32/BN32_010007.cha
./VanDam-Daylong/0metadata.cdc
where /r VanDam-Daylong *
output:
C:\Users\Loann\LAAC\vandam-data\VanDam-Daylong\0metadata.cdc
C:\Users\Loann\LAAC\vandam-data\VanDam-Daylong\BN32\BN32_010007.cha
C:\Users\Loann\LAAC\vandam-data\VanDam-Daylong\BN32\0its\e20100728_143446_003489.its
0metadata.cdc1looks like some metadata file, so we will move it tometadata/:
mv VanDam-Daylong/0metadata.cdc metadata/
move VanDam-Daylong\0metadata.cdc metadata\
BN32_010007.chacontains some transcriptions. Let’s create a set of annotationschaand move it there :
mkdir -p annotations/cha/raw
mv VanDam-Daylong/BN32/BN32_010007.cha annotations/cha/raw
mkdir annotations\cha\raw
move VanDam-Daylong\BN32\BN32_010007.cha annotations\cha\raw
e20100728_143446_003489.itscontains diarization and other information such as word counts. Let’s create another set of annotations for it. And for the sake of consistency, we’ll rename itBN32_010007.its.
mkdir -p annotations/its/raw
mv VanDam-Daylong/BN32/0its/e20100728_143446_003489.its annotations/its/raw/BN32_010007.its
mkdir annotations\its\raw
move VanDam-Daylong/BN32\0its\e20100728_143446_003489.its annotations\its\raw\BN32_010007.its
Now we’ve got all the files. Let’s try to run the validation on the dataset:
child-project validate .
output:
Traceback (most recent call last):
File "/Users/acristia/anaconda3/bin/child-project", line 8, in <module>
sys.exit(main())
File "/Users/acristia/anaconda3/lib/python3.7/site-packages/ChildProject/cmdline.py", line 241, in main
args.func(args)
File "/Users/acristia/anaconda3/lib/python3.7/site-packages/ChildProject/cmdline.py", line 39, in validate
errors, warnings = project.validate(args.ignore_files)
File "/Users/acristia/anaconda3/lib/python3.7/site-packages/ChildProject/projects.py", line 102, in validate
self.read()
File "/Users/acristia/anaconda3/lib/python3.7/site-packages/ChildProject/projects.py", line 86, in read
self.children = self.ct.read(lookup_extensions = ['.csv', '.xls', '.xlsx'])
File "/Users/acristia/anaconda3/lib/python3.7/site-packages/ChildProject/tables.py", line 65, in read
raise Exception("could not find table '{}'".format(self.path))
Exception: could not find table './metadata/children'
child-project validate .
output:
Traceback (most recent call last):
File "C:\Users\acristia\anaconda3\bin\child-project", line 8, in <module>
sys.exit(main())
File "C:\Users\acristia\anaconda3\lib\python3.7\site-packages\ChildProject\cmdline.py", line 241, in main
args.func(args)
File "C:\Users\acristia\anaconda3\lib\python3.7\site-packages\ChildProject\cmdline.py", line 39, in validate
errors, warnings = project.validate(args.ignore_files)
File "C:\Users\acristia\anaconda3\lib\python3.7\site-packages\ChildProject\projects.py", line 102, in validate
self.read()
File "C:\Users\acristia\anaconda3\lib\python3.7\site-packages\ChildProject\projects.py", line 86, in read
self.children = self.ct.read(lookup_extensions = ['.csv', '.xls', '.xlsx'])
File "C:\Users\acristia\anaconda3\lib\python3.7\site-packages\ChildProject\tables.py", line 65, in read
raise Exception("could not find table '{}'".format(self.path))
Exception: could not find table '.\metadata\children'
This is expected. The validation should fail, because the metadata is missing. We need to store the metadata about the children and the recordings in a way that meets the specifications (see Metadata).
Create the metadata
We need two metadata files:
metadata/recordings.csv, which links each recording to their associate metadata(recording date and time, recording device, etc.)
metadata/children.csv, which stores the information about the participants.
Let’s start with the recordings metadata. metadata/recordings.csv
should at least have the following columns: experiment, child_id,
date_iso, start_time, recording_device_type, recording_filename. The .its file
contains (annotations/its/raw/BN32_010007.its) precious information
about when the recording started:
<Recording num="1" startClockTime="2010-07-24T11:58:16Z" endClockTime="2010-07-25T01:59:20Z" startTime="PT0.00S" endTime="PT50464.24S">
The ‘Z’ suffix in the clock times indicate they correspond to the UTC timezone. However, the metadata should contain local times only. The difference between the two is 5 hours, according to the following line in the .its file:
<TransferTime LocalTime="2010-07-28T14:34:46" TimeZone="CST" UTCTime="2010-07-28T19:34:46" />
Therefore, the recording started on 2010-07-24, at 06:58 (local time).
In order to reflect that information, the recordings CSV metadata should look like this (we have decided that the only child of the dataset should have ID ‘1’):
experiment |
child_id |
date_iso |
start_time |
recording_device_type |
recording_filename |
|---|---|---|---|---|---|
vandam-daylong |
1 |
2010-07-24 |
11:58 |
lena |
BN32_010007.mp3 |
We have prepared it for you. Download recordings.csv here,
and save it in the metadata subfolder of your dataset.
You can check its content by issuing the following command:
cat metadata/recordings.csv
output:
experiment,child_id,date_iso,start_time,recording_device_type,recording_filename
vandam-daylong,1,2010-07-24,11:58,lena,BN32_010007.mp3
type metadata\recordings.csv
output:
experiment,child_id,date_iso,start_time,recording_device_type,recording_filename
vandam-daylong,1,2010-07-24,11:58,lena,BN32_010007.mp3
Now, let us proceed to the children metadata. The only fields that are required are: experiment, child_id and child_dob. The .its file also contains some information about the child:
<ChildInfo algorithmAge="P12M" gender="F" />
She was a 12 month old girl at the time of the recording. We can thus
assign her a calculated date of birth: 2009-07-24. We will set
dob_criterion to “extrapolated” to keep track of the fact that the
date of birth was calculated from the approximate age at recording. We
will also set dob_accuracy to ‘month’ for that child.
In other words, the children metadata CSV file should look like this:
experiment |
child_id |
child_dob |
dob_criterion |
dob_accuracy |
|---|---|---|---|---|
vandam-daylong |
1 |
2009-07-24 |
extrapolated |
month |
We have prepared it for you. Download children.csv here,
and save it in the metadata subfolder of your dataset.
You can check its content by issuing the following command:
cat metadata/children.csv
output:
experiment,child_id,child_dob,dob_criterion,dob_accuracy
vandam-daylong,1,2009-07-24,extrapolated,month
type metadata\children.csv
output:
experiment,child_id,child_dob,dob_criterion,dob_accuracy
vandam-daylong,1,2009-07-24,extrapolated,month
We can now make sure that they are no errors by running the validation command again:
child-project validate .
child-project validate .
No error occurs.
Note
The metadata can be enriched with many more columns. See Metadata for standard columns. You can add as many extra, custom columns as you need.
Save the changes locally
A DataLad dataset is essentially a git repository, with the large files being handled by git-annex. Some of the files (usually the small, text files such as metadata and scripts) ought to be versionned with git, and the larger files or binary files should be stored in the annex.
The rules to decide what files should be stored which way can be set in
the .gitattributes file. You should fill it by running the following
lines:
echo "* annex.backend=MD5E" >.gitattributes
echo "**/.git* annex.largefiles=nothing" >>.gitattributes
echo "scripts/* annex.largefiles=nothing" >>.gitattributes
echo "metadata/* annex.largefiles=nothing" >>.gitattributes
echo "recordings/converted/* annex.largefiles=((mimeencoding=binary))" >>.gitattributes
echo "* annex.backend=MD5E" >.gitattributes
echo "**/.git* annex.largefiles=nothing" >>.gitattributes
echo "scripts/* annex.largefiles=nothing" >>.gitattributes
echo "metadata/* annex.largefiles=nothing" >>.gitattributes
echo "recordings/converted/* annex.largefiles=((mimeencoding=binary))" >>.gitattributes
Check the content by running:
cat .gitattributes
output:
* annex.backend=MD5E
**/.git* annex.largefiles=nothing
scripts/* annex.largefiles=nothing
metadata/* annex.largefiles=nothing
recordings/converted/* annex.largefiles=((mimeencoding=binary))
type .gitattributes
output:
* annex.backend=MD5E
**/.git* annex.largefiles=nothing
scripts/* annex.largefiles=nothing
metadata/* annex.largefiles=nothing
recordings/converted/* annex.largefiles=((mimeencoding=binary))
These rules will version all the files under scripts/ and
metadata/, as well as the text files inside of
recordings/converted/. By default, the other files will be put in
the annex.
The changes can now be saved. This can be done with datalad
save.
datalad save is equivalent to a combination of git add and
git commit in one go. It decides, based on the rules in
.gitattributes, whether to store files with git or git-annex.
datalad save . -m "first commit"
datalad save . -m "first commit"
However, so far, your changes remain local, and your dataset still needs to be published into a sibling to be shared with others.
Processing
You can do some processing on the dataset. For instance, you can compute the duration of the recording, and update the metadata with this information. This is easily done with:
child-project compute-durations .
child-project compute-durations .
Warning
We are currently aware of a problem with the compute-duration command on Windows systems, it is likely this will fail for now.
Now metadata/recordings.csv became:
cat metadata/recordings.csv
output:
experiment,child_id,date_iso,start_time,recording_device_type,recording_filename,duration
vandam-daylong,1,2010-07-24,11:58,lena,BN32_010007.mp3,50464512
type metadata\recordings.csv
output:
experiment,child_id,date_iso,start_time,recording_device_type,recording_filename,duration
vandam-daylong,1,2010-07-24,11:58,lena,BN32_010007.mp3,50464512
You can also convert and index the its annotation:
child-project import-annotations . --set its \
--recording_filename BN32_010007.mp3 \
--time_seek 0 \
--range_onset 0 \
--range_offset 50464512 \
--raw_filename BN32_010007.its \
--format its
child-project import-annotations . --set its ^
--recording_filename BN32_010007.mp3 ^
--time_seek 0 ^
--range_onset 0 ^
--range_offset 50464512 ^
--raw_filename BN32_010007.its ^
--format its
And save the changes again:
datalad save . -m "its"
datalad save . -m "its"
Publish the dataset
Where to publish my dataset ?
DataLad allows you to publish your datasets on large number of storage providers, including Amazon S3, Dropbox, Google Cloud Storage, Microsoft Azure Blob Storage, etc., each having their own advantages and limitations. It is also possible to publish to several platforms, as we do with our own datasets.
The table below summarises the features of a few storage supports. The solutions described here are by no mean exhaustive, but they are easy to generalize.
Platforms that support Git store the .git files and will allow you to clone the datasets from them with
datalad installPlatforms that support Large Files will allow you to store and distribute the large or binary files that are stored with git-annex instead of the regular git files (such as scripts and metadata)
It is necessary to use a platform or a combination of platforms that supports both. We recommend the use of GIN, although you should always push your data to another platform as backup.
Provider |
Git |
Large Files |
Authentication |
Permissions |
Cost |
Quota |
|---|---|---|---|---|---|---|
GIN |
Yes |
Yes |
HTTPS/SSH |
ACL |
Free below ~10 TB |
None |
SSH server |
Yes |
Yes |
SSH |
Unix |
- |
None |
GitHub |
Yes |
No |
HTTPS/SSH |
ACL |
Free |
~1 GB |
GitLab |
Yes |
No |
HTTPS/SSH |
ACL |
Free |
~1 GB |
Amazon S3 |
No |
Yes |
API |
IAM |
~4$/TB/month |
None |
Nextcloud |
No |
Yes |
WebDav |
ACL |
- |
None |
OSF.io |
Yes |
Yes* |
Token |
ACL |
Free |
5 GB |
Note
DataLad uses git-annex, which naturally handles encryption. This is particularly useful when using third-party providers such as Amazon S3.
Publish to GIN
Note
Before anything, you will need to create an account on GIN, and to link your SSH public key to your GIN account.
Create a new repository from GIN’s web interface. Uncheck “Initialize this repository with selected files and template”
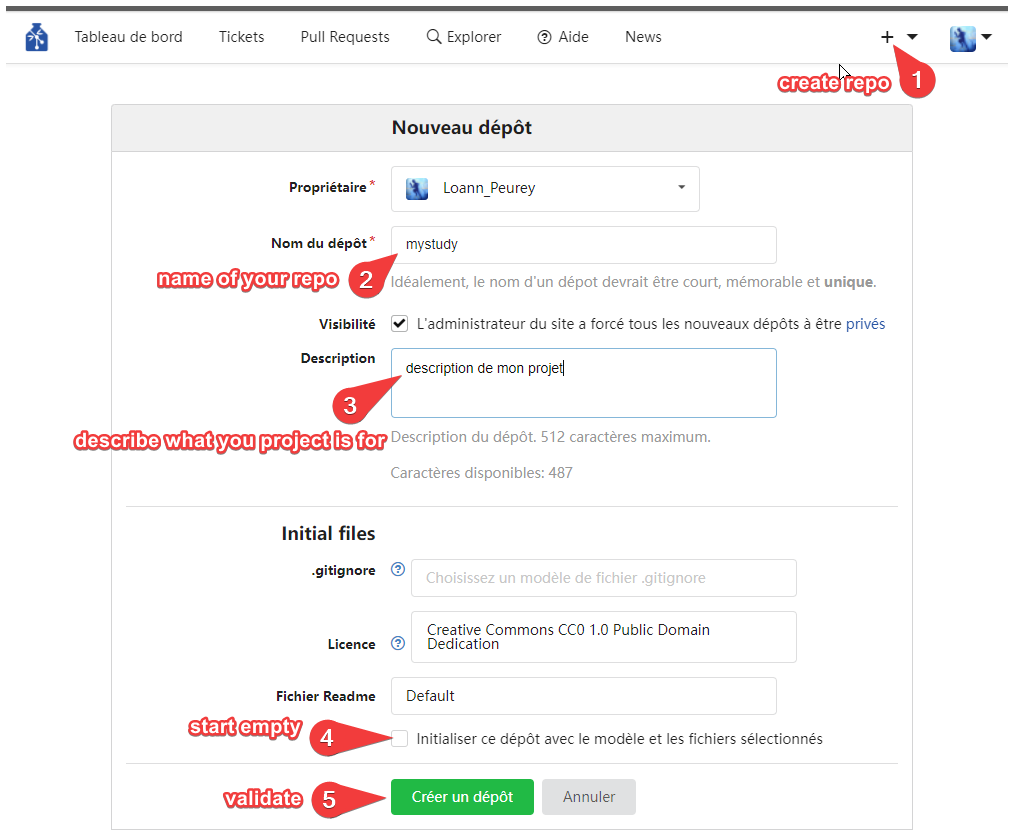
creating a repository on GIN
Copy the SSH url of your repository to your clipboard, e.g.:
git@gin.g-node.org:/<username>/<reponame>.gitAdd a datalad sibling pointing to this repository:
datalad siblings add \
--name gin \
--url git@gin.g-node.org:/<username>/<reponame>.git
Push the data to GIN:
datalad push --to gin
Publish to a SSH server
If you have access to a SSH server with enough storage capacity, you can use it to store and share the dataset. This is done with the datalad create-sibling command:
datalad create-sibling [-h] [-s [NAME]] [--target-dir PATH] [--target-url URL] [--target-pushurl URL] [--dataset DATASET] [-r] [-R LEVELS] [--existing MODE] [--shared {false|true|umask|group|all|world|everybody|0xxx}] [--group GROUP] [--ui {false|true|html_filename}] [--as-common-datasrc NAME] [--publish-by-default REFSPEC] [--publish-depends SIBLINGNAME] [--annex-wanted EXPR] [--annex-group EXPR] [--annex-groupwanted EXPR] [--inherit] [--since SINCE] [SSHURL]
For instance, you can create it (this is only to be done once) by issuing:
datalad create-sibling -s cluster --annex-wanted 'include=*' <ssh-server>:/remote/path/to/the/dataset
cluster is the name of the sibling, and
<ssh-server>:/remote/path/to/the/dataset is the SSH url of its
destination. --annex-wanted 'include=*' implies that all large files
will be published to this sibling by default.
Once the sibling has been created, the changes can be published:
datalad push --to cluster
That’s it! People can now get your data from:
datalad install <ssh-server>:/remote/path/to/the/dataset
If --annex-wanted had not been set to 'include=*', the large
files (i.e. annexed files) would not be published unless you asked for
it explicitly with the --data flag:
datalad push --to cluster --data anything
Publish to GitHub
You first need to create the repository, which can be done in a straightforward way from the command line with datalad create-sibling-github:
datalad create-sibling-github [-h] [--dataset DATASET] [-r] [-R LEVELS] [-s NAME] [--existing MODE] [--github-login NAME] [--github-organization NAME] [--access-protocol {https|ssh}] [--publish-depends SIBLINGNAME] [--private] [--dryrun] REPONAME
For instance:
datalad create-sibling-github -s origin --access-protocol ssh vandam-daylong-demo
origin will be the local name of the sibling, and
vandam-daylong-demo the name of the GitHub repository. Once the
sibling has been created, you can publish the changes with datalad
push:
datalad push --to origin
You should get a repository identical to this one.
Users can now install your dataset from GitHub:
datalad install https://github.com/LAAC-LSCP/vandam-daylong-demo.git
PS: we recommend that you do git push --set-upstream origin to set
upstream to the GitHub sibling. Users who install your dataset from GitHub will not
need to do this.
GitHub + SSH mirror to store the large files
Now, let’s assume you have already created a SSH sibling as well for
your dataset, and that it is named cluster. You can make sure that
all changes to github are published to cluster as well, by
setting the publish-depends property of the github sibling:
datalad siblings configure -s origin --publish-depends cluster
Now, datalad push --to origin will publish the changes to both
cluster and github.
However, when the users install your dataset from GitHub, they will not
have access to the cluster sibling unless you make it available to
them, which can be done this way :
git annex
git annex initremote cluster type=git location=ssh://cluster.com/path/to/the/repository autoenable=true
git annex enableremote cluster
git remote add origin git@github.com:LAAC-LSCP/vandam-daylong-demo.git
Publish on S3
Like other git annex special remotes, Amazon S3 will not support the git files, only the large files. It could be used together win GitHub as the primary host for your large files, or as a backup.
For the sake of simplicity, we will not use encryption here, but git annex implements severalencryption schemeswhich are easy to use.
First, store your AWS credentiels into your environment variables, like this:
export AWS_ACCESS_KEY_ID="08TJMT99S3511WOZEP91"
export AWS_SECRET_ACCESS_KEY="s3kr1t"
You are now readyto create the s3 sibling. This is done directly through git-annex this time:
git annex initremote s3 chunk=100MiB type=S3 encryption=none datacenter=eu-west-3 embedcreds=no signature=v4
You can now publish the data with:
datalad push --to s3 --data anything
(Optional) You can set the S3 sibling to require that all large files should be stored on it:
datalad siblings configure -s s3 --annex-wanted 'include=*'
This will let DataLad publish all the large files automatically without
setting --data:
datalad push --to s3
Let’s assume your users will install the dataset from a GitHub repository. You should publish the information about the newly created S3 sibling on GitHub, which can be done with (provided you have set up your GitHub repository as described in the previous section):
datalad push --to github
Now, users will be able to get the data by issuing the following commands:
datalad install git@github.com:<your-username>/vandam-daylong-demo.git
git annex enableremote s3
datalad get *
With this configuration, they will need to setup their AWS credentials as you did. But it is possible to configure the sibling so that the credentials are encrypted and stored in the repository, so all users with authorized private keys will be able to get the data without this step.
Publish on OSF
DataLad has an extension to publish data on the Open Science Framework.
This extension supports the following modes:
Mode |
datalad install |
large files |
history |
older files |
human-readable project |
|---|---|---|---|---|---|
|
Yes |
Yes |
Yes |
Yes |
No |
|
Yes |
Yes |
Yes |
No |
Yes |
|
Yes |
No |
Yes |
No |
No |
|
No |
Yes |
No |
Yes |
Yes |
The first step is to install the extension:
pip install datalad-osf --upgrade
We decide to use the export mode - but you can decide which best
suits your needs from the table above. We can now create the sibling:
datalad create-sibling-osf --title "VanDam Demo" \
--mode export \
-s osf \
--category data \
--tag reproducibility \
--public
You will be prompted your credentials in the process, which will require access tokens to be created from your osf.io account.
And finally we can push the data. This is done in two steps:
publishing the .git files so people can clone the dataset directly from OSF
datalad push --to osf
exporting a human-readable snapshot of the files to OSF
git-annex export HEAD --to osf-storage